NYU Langone scientists find a new way to outsmart deadly bacteria that have become resistant to antibiotics and kill thousands each year.
Photo: Callista Images/Getty
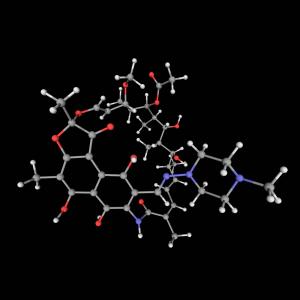
Ever taste something awful and instinctively spit it out? The deadly bacterium Staphylococcus aureus relies on a similar instinct, using a pumping mechanism to expel antibiotics that could kill it. It’s just one clever way that S. aureus has evolved over the years to outsmart more than 60 common antibiotics, intensifying a global crisis of antibiotic-resistant infections that claim some 700,000 lives each year.
Now, researchers at NYU Grossman School of Medicine and NYU Langone’s Perlmutter Cancer Center have unlocked the mysteries of a bacterial mechanism that science has long sought to solve, and discovered a potential way to disarm this so-called “efflux pump.” In a paper published in Nature Chemical Biology, the researchers developed a clever strategy to visualize the infinitesimally small parts of the pump and, in the process, engineered an antibody that could jam it. In cell cultures, a protein fragment of the antibody reduced the growth of antibiotic-resistant S. aureus by more than 95 percent at high concentrations when combined with the antibiotic norfloxacin.
“Instead of trying to find a new antibiotic, we aimed to make commonly used antibiotics that have been rendered ineffective by bacterial resistance highly effective again,” says study author Douglas Brawley, PhD, who completed his doctoral thesis in the laboratories of fellow study authors Nathaniel J. Traaseth, PhD, professor in NYU’s Department of Chemistry, and Da-Neng Wang, PhD, professor in the Department of Cell Biology at NYU Grossman School of Medicine.
This work is particularly striking for its collaborative effort, drawing upon experts in structural biology, antibody engineering, microbiology, and peptide chemistry. “The discovery of this new way to inhibit resistant strains of S. aureus demonstrates that five labs from four departments can collaborate to accomplish what none could alone,” says study author Shohei Koide, PhD, professor in the Department of Biochemistry and Molecular Pharmacology at NYU Grossman School of Medicine.