Rheumatologist Dr. Jose U. Scher leads teams that study drug responses in rheumatoid arthritis patients, as well as predictions for psoriatic arthritis patients based on their symptom progression.
Photo: NYU Langone staff
With growing awareness of the gut microbiome’s key role in mediating sickness and health, scientists are ramping up investigations of whether the pathogenic mechanisms of certain microbes may trigger rheumatoid arthritis (RA) and other autoimmune diseases. NYU Langone Health researchers are conducting a related but novel assessment of how gut flora may be altering the efficacy and toxicity of autoimmune disease–targeting medications. Additionally, a study is underway that could ultimately help clinicians predict the psoriasis patients at highest risk for psoriatic arthritis.
Bacterial Enzymes May Decrease Methotrexate Absorption
In the first set of studies, a team led by Jose U. Scher, MD, associate professor in the Department of Medicine and director of the Psoriatic Arthritis Center and the Microbiome Center for Rheumatology and Autoimmunity, is asking whether gut flora–produced enzymes may metabolize oral methotrexate and decrease its intestinal absorption, thereby reducing its effectiveness in RA patients. The question is critical for clinical care, given that methotrexate remains the anchor drug for treating RA. The problem is that more than half of patients with moderate or severe arthritis show either no symptom improvement in response to this medication or, at best, suboptimal improvement.
“What we’re seeing is that for some patients, certain genes within the gut flora may predict whether or not they’re going to respond to the methotrexate,” Dr. Scher says.
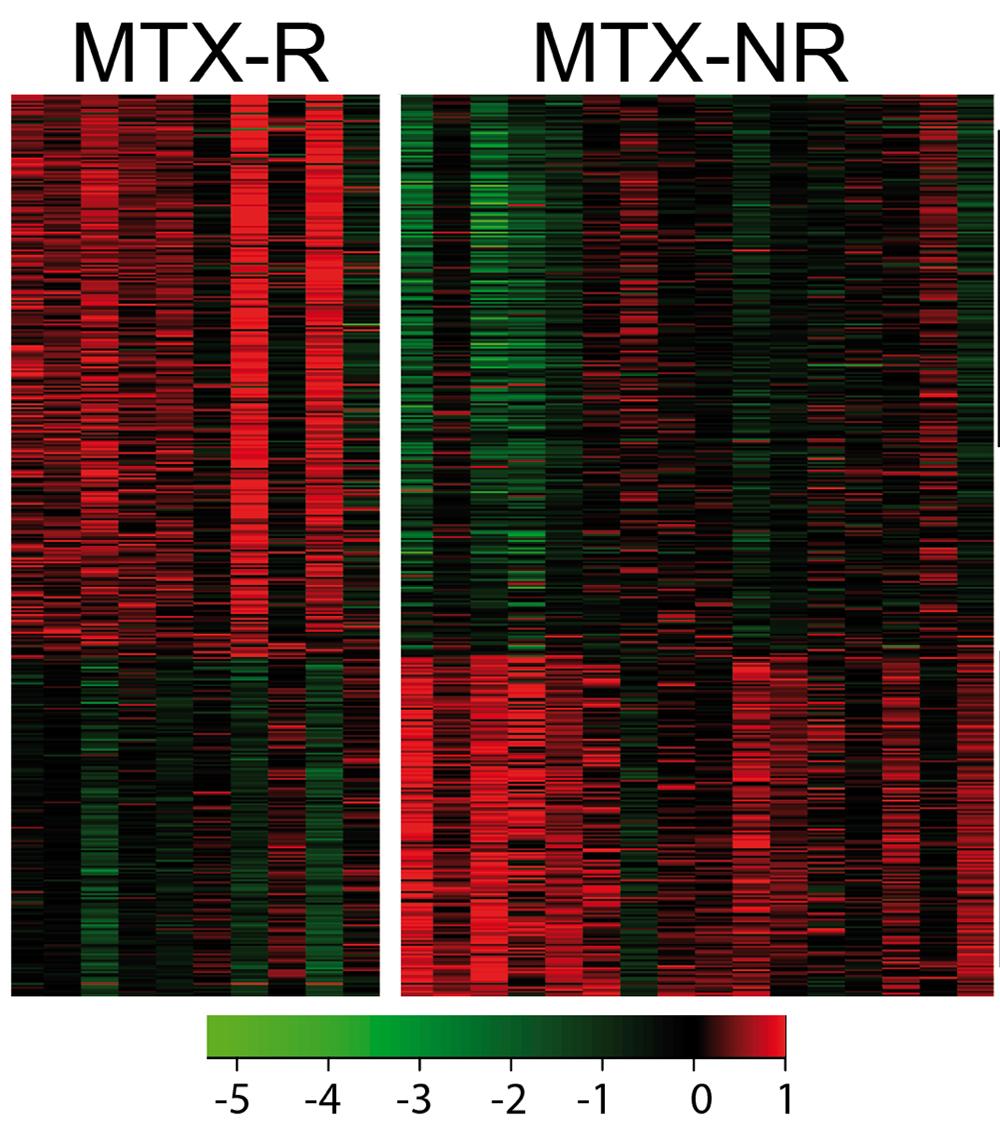
Several National Institutes of Health (NIH) grants, including a prestigious new R01 award, are helping the researchers address this critical clinical question. For one set of ex vivo experiments, Dr. Scher’s team took gut flora cultures from the baseline fecal samples of new-onset patients whose RA subsequently responded or failed to respond adequately to methotrexate.
When researchers added the cultures to Petri dishes containing the drug, they saw significant variability in the amount of methotrexate that remained in the dish two to three days later. Importantly, Dr. Scher says, “there is a correlation between the methotrexate that disappears, or gets metabolized, and the corresponding patient’s response to the drug in the real world.”
Metabolism Study and Machine Learning Implicate Multiple Genes
Dr. Scher and other researchers hypothesize that when more methotrexate is metabolized in a patient’s intestinal lumen, less of it is absorbed and available to help fight the disease. Instead of specific microbes mediating the metabolism, he suspects that similar enzyme-encoding genes from multiple species may be collectively responsible. So far, his research has identified dozens of microbial genes, some of which have been directly implicated in methotrexate metabolism. The role of others remains unknown.
Simultaneously, Dr. Scher and collaborators are feeding clinical phenotypic, metagenomic, metabolomic, and other data into machine learning algorithms. The methods can help generate predictions about which genes and mechanisms are most relevant in driving in vivo drug alterations. Using an algorithm called “random forest,” for example, the team identified 39 microbial genes that predict a methotrexate response.
Advancing Toward Microbiome-Mediated Precision Medicine
If supported by further research, Dr. Scher’s finding might help advance precision medicine via analysis of individual patients’ gut microbiomes: the relative abundance of specific microbial genes might yield a signature that warns rheumatologists that a different therapeutic strategy may be necessary. For patients whose gut flora are more adept at metabolizing methotrexate, doctors might consider administering the drug subcutaneously or switching to a different medication.
Alternatively, ongoing research with collaborators at the University of California, San Francisco, is trying to optimize methotrexate through modification experiments in mice. “If you colonize the mice with different human microbiomes, can you modify them in such a way that they no longer metabolize methotrexate?” Dr. Scher asks. Adding various probiotics or prebiotics, for example, may decrease the efficiency with which a microbiome metabolizes the drug, thereby improving the drug’s bioavailability for ameliorating RA symptoms.
Together, these lines of research could eventually lead to markedly improved outcomes for patients whose own gut flora may be thwarting an effective drug response.
From Psoriasis to Psoriatic Arthritis: Predicting the Progression
Nearly one third of patients with psoriasis will develop psoriatic arthritis. As a part of a suite of studies known as Preventing Psoriatic Arthritis, or PreP, Dr. Scher and colleagues have launched two new efforts to better predict the progressing patients.
In July 2019, the National Psoriasis Foundation awarded Rebecca Haberman, MD, a clinical instructor in the Department of Medicine, a prestigious early career research grant. Dr. Haberman, also one of NYU Langone’s recipients of an NIH T32 training award, is leading a study to compare four patient populations at different stages and risk for psoriatic arthritis development. The goal is to determine the most relevant clinical, genetic, and microbiome-based risk factors of disease progression.
Dr. Haberman’s aim is a predictive model that could help clinicians and scientists accurately assess the risk of psoriatic arthritis development in psoriasis patients. In turn, the work could accelerate early diagnoses and inform preventive clinical trials by identifying the patients at highest risk.
Spotting Psoriatic Arthritis Symptoms with a Smartphone App
A separate multicenter collaboration called the Psorcast Study is aimed at developing digital biomarkers of psoriatic arthritis based on smartphone sensors. “Essentially, the study is asking how we can obtain data that is passively captured through gyroscopes and accelerometers that live in our smartphones,” Dr. Scher says.
The researchers are asking psoriasis patients to pay attention to digital markers that may highlight telltale symptoms and provide a personalized forecast of psoriatic arthritis risk. For this proof-of-concept study, patients use a phone app to assess their joint function via gyroscope and accelerometer data, take photos of joint swelling and psoriasis plaques, and record other patient-reported outcomes.
The study, done in collaboration with Psoriasis and Psoriatic Arthritis Clinics Multicenter Advancement Network (PPACMAN) and Sage Bionetworks, could lead to an open-source repository of patient-captured data for future research and eventually to a multifaceted diagnostic test with the promise of detecting the earliest signs of disease.