Credit: Getty/Visoot Uthairam
A computer program based on data from nearly a half-million tissue images and powered by artificial intelligence (AI) can accurately diagnose in cases of adenocarcinoma, the most common form of lung cancer, a new study shows.
Researchers at NYU Langone Health’s Perlmutter Cancer Center and the University of Glasgow, in Scotland, developed and tested the program. They say that because it incorporates structural features of tumors from 452 adenocarcinoma patients, who are among the more than 11,000 patients in the U.S. National Cancer Institute’s Cancer Genome Atlas, the program offers an unbiased, detailed, and reliable second opinion for patients and oncologists about the presence of the cancer and the likelihood and timing of its return, also known as its prognosis.
The research team also points out that the program is independent and “self-taught,” meaning that it determined on its own which structural features were statistically most significant for gauging the severity of disease and had the greatest impact on tumor recurrence.
Published online June 11 in the journal Nature Communications, the study program, also called an algorithm, or more specifically histomorphological phenotype learning (HPL), was found to accurately distinguish between similar lung cancers—adenocarcinoma and squamous cell cancers—99 percent of the time. The HPL program was also found to be 72 percent accurate at predicting the likelihood and timing of cancer’s return after therapy, bettering the 64 percent accuracy in the predictions made by pathologists who directly examined the same patients’ tumor images, researchers say.
“Our new histomorphological phenotype learning program has the potential to offer cancer specialists and their patients a quick and unbiased diagnostic tool for lung adenocarcinoma that, once further testing is complete, can also be used to help validate and even guide their treatment decisions,” said study lead investigator Nicolas Coudray, PhD, a bioinformatics programmer at NYU Grossman School of Medicine and Perlmutter Cancer Center.
“Patients, physicians, and researchers know they can rely on this predictive modeling because it is self-taught, provides explainable decisions, and is based only on the knowledge drawn specifically from each patient’s tissue, including such features as its proportion of dying cells and tumor-fighting immune cells and how densely packed the tumor cells are,” said Dr. Coudray.
“Lung tissue samples can now be analyzed in minutes by our computer program to provide fairly accurate predictions of whether their cancer will return, predictions that are better than current standards of care for making a prognosis in lung adenocarcinoma,” said study co-senior investigator Aristotelis Tsirigos, PhD. Dr. Tsirigos is a professor in the Departments of Pathology and Medicine at NYU Grossman School of Medicine and a member of Perlmutter Cancer Center; he also serves as co-director of precision medicine and director of NYU Langone’s Applied Bioinformatics Laboratories.
Dr. Tsirigos says that thanks to such tools and other advances in learning about the biology of lung cancer, pathologists will examine tissue more often through scans on their computer screens and less via microscopes, and then use their AI program to analyze the image and produce its own image of the scan.
The new image, or “landscape,” they add, will offer a detailed breakdown of the tissue’s content. It might note, for example, that there is 5 percent necrosis and 10 percent tumor infiltration and what that means in terms of survival. That reading may statistically equate to an 80 percent chance of remaining cancer-free for two years or more, based on information from all the patient data in the program.
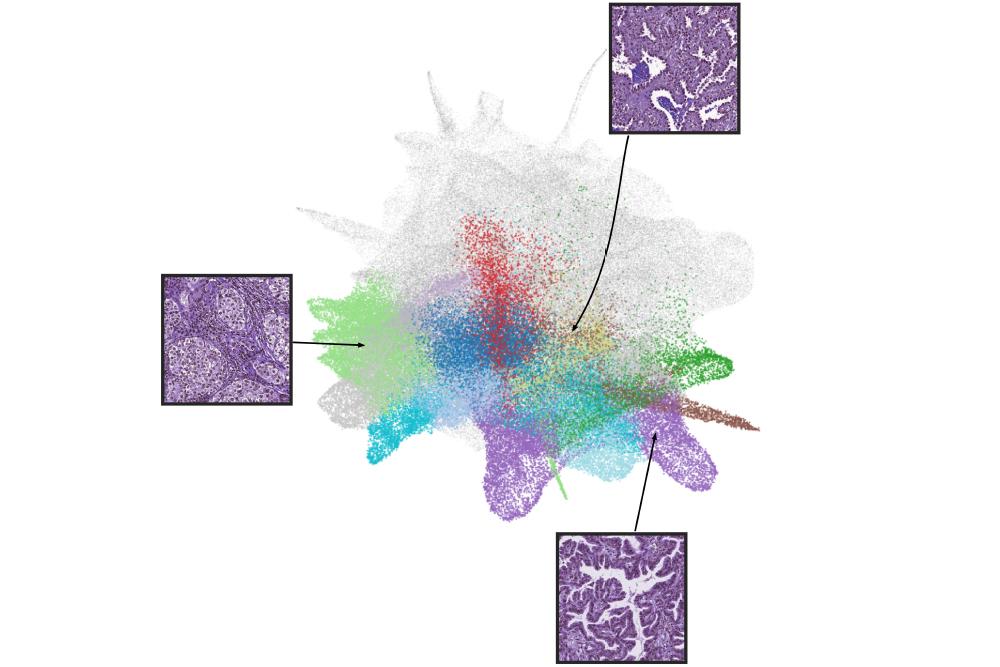
To develop the HPL program, the researchers first analyzed lung adenocarcinoma tissue slides from the Cancer Genome Atlas. Adenocarcinoma was chosen for the test model because the disease is known for characteristic features. As an example, they note that its tumor cells tend to group in so-called acinar—saclike patterns—and spread predictably along the surface lining of lung cells.
From their analysis of the slides, whose visual images were digitally scanned and broken into 432,231 small quadrants or tiles, researchers found 46 key characteristics, which they term histomorphological phenotype clusters, from both normal and diseased tissue. A subset of these clusters was statistically linked to either cancer’s early return or to long-term survival. The findings were then confirmed by further and separate testing on tissue images from 276 men and women who were treated for adenocarcinoma at NYU Langone from 2006 to 2021.
Researchers say their goal is to use the HPL algorithm to assign to each patient a score between 0 and 1 that reflects their statistical chance of survival and tumor recurrence for up to five years. They stress that because HPL is self-learning, the program will become increasingly more accurate as more data is added over time. To build public trust, researchers have posted their programming code online and have plans to make the new HPL tool freely available upon completion of further testing.
Characteristics linked to tumors recurring included high tile percentages of dead cancer cells and tumor-fighting immune cells called lymphocytes, and the dense clustering of tumor cells in the outer linings of the lungs. Features tied to increased likelihood for survival were high percentages of unchanged or preserved lung sac tissue, and the lack of, or the mild presence of, inflammatory cells.
Dr. Tsirigos says the team next plans to look at developing HPL-like programs for other cancers, such as breast, ovarian, and colorectal, that are similarly based on distinctive and key morphological features and additional molecular data. The team also has plans to expand and improve the accuracy of the current adenocarcinoma HPL program by including other data from hospital electronic health records about other illnesses and diseases, or even income and home zip code.
Funding support for the new study was provided by National Institutes of Health grant P30CA016087, United Kingdom Research Council grants Ep/R018634/1 and BB/V016067/1, and European Union Horizon 2020 grant 101016851.
Besides Dr. Tsirigos and Dr. Coudray, other NYU Langone researchers involved in this study are Anna Yeaton, Bojing Liu, Hortense Le, Luis A. Chiriboga, Afreen Karimkhan, Navneet Narula, Christopher Y. Park, Harvey I. Pass, and Andre L. Moreira. Study co-lead investigator Adalberto Claudio Quiros, study co-investigators Xinyu Yang and John Le Quesne, and study co-senior investigator Ke Yuan are all at the University of Glasgow. Study co-investigator David Moore is at University College London.
Media Inquiries
David March
Phone: 212-404-3528
David.March@NYULangone.org