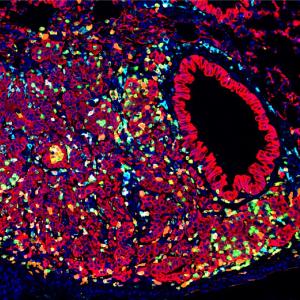
Proteins that assemble into the machinery needed for DNA repair in live bacterial cells.
Image: Bharati et al., Nature
Two recent studies provide a radically new picture of how bacterial cells continually repair damaged sections (lesions) in their DNA.
Led by researchers from NYU Grossman School of Medicine, the work revolves around the delicacy of DNA molecules, which are vulnerable to damage by reactive byproducts of cellular metabolism, toxins, and ultraviolet light. Given that damaged DNA can result in detrimental DNA code changes (mutations) and death, cells evolved to have DNA repair machineries. A major unresolved question in the field, however, is how do these machineries rapidly search for and find rare stretches of damage amid the “vast fields” of undamaged DNA.
Past studies had found that one important search mechanism—transcription-coupled repair, or TCR—relies on RNA polymerase, the large protein machine (complex) that motors down the DNA chain, reading the code of DNA “letters” as it transcribes instructions into RNA molecules, which then direct protein building. Going into the current study, however, the TCR mechanism was misunderstood, say the study authors.
Widely accepted work, including studies that led to a 2015 Noble Prize in Chemistry, had argued that TCR played a relatively small role in repair because it relied on a putative TCR factor that made only a marginal contribution to DNA repair. A parallel process, global genome repair (GGR), was assumed to scan and fix most of DNA independent of transcription. Both processes were thought to set the stage for nucleotide excision repair (NER), in which a damaged stretch of DNA was snipped out and replaced by an accurate copy.
Now two new studies published online March 30 in Nature and Nature Communications agree, based on the first-of-its kind, multistage analysis of DNA repair in living Escherichia coli cells, that most, if not all, NER is coupled to RNA polymerase, which scans the entire bacterial genetic code for damage.
“Based on our results, we need to rethink some of the basic theories in the DNA repair field,” says senior study author Evgeny A. Nudler, PhD, the Julie Wilson Anderson Professor of Biochemistry in the Department of Biochemistry and Molecular Pharmacology at NYU Langone Health. “A true understanding of such repair is a fundamental goal in medicine, as most antibiotics and chemotherapies kill disease-causing cells by damaging their DNA, and the ability to halt repairs would make such cells much more vulnerable to existing drugs,” adds Dr. Nudler, who is also an investigator with the Howard Hughes Medical Institute.
Discovery Pipeline
Past studies could not fully capture the biological reality of NER in bacteria, say the current authors, because they used experiments that tried to recreate complex protein interactions outside of living cells. That led the field, for instance, to define a protein called Mfd as the central player in TCR, even as most DNA repair was found to proceed whether or not Mfd was present. This, in turn, suggested that TCR was a minor repair pathway. TCR was also thought to happen only within the DNA regions that are highly transcribed. Seldom-transcribed genomic locations, or parts of the genome assumed to be “non-transcribed,” were thought to be subject to GGR.
The study newly published in Nature used a groundbreaking technology called crosslinking mass spectrometry (XLMS) to map the distances between chemically linked proteins, and so determine the interacting surfaces of massive NER and polymerase complexes for the first time as they are assembled in living cells. The team then fed the spectrometry data into computer-driven simulations, culminating in realistic structural models.
Contrary to the conventional dogma, the study found that RNA polymerase serves as the scaffold for the assembly of the entire NER complex, and as the primary sensor of DNA lesions. It turned out that the principal NER enzymes UvrA and UvrB do not locate most lesions on their own, but are delivered to them by RNA polymerase. This fundamental TCR process is independent of Mfd, say the authors.
The second study, published in Nature Communications, again in living cells, used a high-throughput sequencing technology called cyclobutane pyrimidine dimer sequencing (CPD-seq) to track the appearance of DNA lesions upon exposure to ultraviolet light, and the rate of repair with a resolution down to a single letter (nucleotide) in the DNA code. CPD-seq showed that interfering with bacterial transcription using the antibiotic rifampicin shuts down repair throughout the bacterial genome. The study findings argue that NER is tightly coupled to transcription everywhere in the bacterial chromosome, the DNA infrastructure that houses all the genes.
In another fascinating leap, experiments showed that bacterial cells, in the face of DNA damage, inhibit the action of the protein Rho, the global termination signal which tells RNA polymerase to stop reading. With the stop signals dialed down, RNA polymerases read on and on, delivering the repair enzymes to DNA damage anywhere it was encountered throughout the genome.
“Given our findings, we theorize that eukaryotes, including human cells, also use RNA polymerase for efficient repair globally, as the bacterial TCR complexes described here have human analogs,” says co-first author of the Nature study Binod Bharati, PhD, a postdoctoral scholar in Dr. Nudler’s lab. “Moving forward, our team plans to confirm the presence of global TCR in human cells, and if confirmed, to explore whether in the future repair might be safely boosted to counter diseases of aging.”
Study Authors and Funding
Along with Dr. Nudler and Dr. Bharati, the authors of the study published in Nature from the Department of Biochemistry and Molecular Pharmacology are co-first study author Manjunath Gowder, Khaled Alzoubi, Vladimir Svetlov, Venu Kamarthapu, Jacob Weaver, Vitaliy N. Epshtein, and Nikita Vasilyev. Additional authors were Fangfang Zheng, Liqiang Shen, and Yu Zhang of the Key Laboratory of Synthetic Biology, Chinese Academy of Sciences Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology of the Chinese Academy of Sciences in Shanghai, China. This work was supported by National Institutes of Health (NIH) grant R01 GM126891, National Key Research and Development Program of China grant 2018YFA0903701, Strategic Priority Research Program of the Chinese Academy of Sciences grant XDB29020302, Chinese Natural Science Foundation of China grant 31822001, and Shanghai Science and Technology Innovation Program grant 19JC1415900.
The first author of the Nature Communications study from the Department of Biochemistry and Molecular Pharmacology was Britney Martinez. Additional authors of this study were Dr. Nudler, Dr. Bharati, and Dr. Epshtein. The work in this paper was supported by NIH grants F31 GM131516-02 and R01 GM126891.
Both studies were supported by the Blavatnik Family Foundation and the Howard Hughes Medical Institute.
Media Inquiries
Greg Williams
Phone: 212-404-3500
gregory.williams@nyulangone.org