Photo: stock_colors/Getty
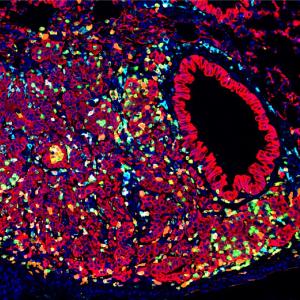
Three-dimensional, lab-grown “mini-corneas” resemble the developing human cornea, making them a powerful new tool for the study of corneal diseases, a study finds.
Led by researchers at NYU Langone Health, the work involves the cornea, the outermost, transparent eye layer that bends light onto the retina so the brain can process images. Cornea disorders are the third leading cause of blindness, and transplant of donor corneas is often the only available treatment option. More than 12 million people are estimated to need cornea transplants, an unmet need that is driving research into cell-based regenerative therapies.
Published in PNAS Nexus, the findings promise to advance the field’s understanding of the molecular events involved in human corneal development in the womb, along with efforts to develop improved therapies for disorders of the cornea, say the study authors. In the future, these organoids could be the source of cells used in cell therapies for corneal diseases.
To do the study, the research team designed corneal organoids, which unlike traditional cell cultures containing a sheet of one type of cells in a dish of nutrients, instead grow stem cells with specific nutrient adjustments such that multiple cell types develop and become organized in three dimensions. With many cell types interacting in layers, the technology more closely mimics the structure and function of the tissues they model than older methods.
“Our study is the first to examine human corneal organoids at a single-cell resolution,” says corresponding study author Shukti Chakravarti, PhD, professor in the Departments of Ophthalmology and Pathology at NYU Grossman School of Medicine. “By reading the genetic code built by active genes in this model, we have found that our organoids behave like actual corneas that are rapidly maturing in the womb.”
The study result revolves around gene expression, wherein molecular instructions encoded in the DNA of activated genes, are converted into related molecules called messenger RNA in real time, on the way to building the proteins that let cells function as part of tissues. The NYU Langone team used an approach called single-cell RNA sequencing to determine cell-specific genes activated in lab-derived human corneal organoids and in corneas from three different adult cadaver human corneas.
Single-cell RNA sequencing revealed that the adult corneas were made up mostly of epithelial cells (outer layer) and stromal cells (middle layer), along with a few cells representing the inner-most endothelium. The study organoids, however, were found to contain an abundance of cells in all three layers. In all, one-third to one-fourth of the developing cells in the organoids had an endothelial signature. And interestingly, the activated gene signature of the organoids resembled that of a developing immature cornea.
“These organoids provide an opportunity to examine gene expression during development,” said Dr. Chakravarti. “With their 3D structures and coexisting cell types they allow the study of cellular signaling and cell–cell interactions in a more natural environment.”
In addition to understanding human cornea development, the study organoids will provide a system for screening for potential therapies for genetic diseases in the future, at less cost than traditional mouse model studies.
Along with Dr. Chakravarti, other NYU Langone authors were George Maiti, PhD, and Nan Hu, MS, in the Department of Ophthalmology; Igor Dolgalev, PhD, in the Applied Bioinformatics Laboratories; and Aristotelis Tsirigos, PhD, in the Applied Bioinformatics Laboratories and the Departments of Medicine and Pathology. Other authors were Maithê Rocha Monteiro de Barros, DVM, PhD, at Columbia University; Mona Roshan, MD, and Karl J. Wahlin, PhD, at the University of California, San Diego; and James W. Foster, PhD, at Johns Hopkins School of Medicine. Funding for the study was provided by National Institutes of Health grants R01EY030917 and R01EY026104.
Media Inquiries
Greg Williams
Phone: 212-404-3500
gregory.williams@nyulangone.org